name: title layout: true class: center, middle, style1 --- # THE TITLE IS OBVIOUSLY MISSING <br> ### [Kevin Cazelles](http://kevincazelles.fr), [Universty of Guelph](https://www.uoguelph.ca/) [](https://github.com/KevCaz/seminarInteractionsDistributions) <br/> ### Département de biologie, Université de Sherbrooke #### October, 12<sup>th</sup> 2018 --- name: style1 layout: true class: style1 --- # ABOUT ME <hr> ## Theory - biogeography - co-occurrence - food webs - population dynamics - homogenezation -- ## Statistics, algorithms, data, software .right[<img src="img/logo/inSilecoLogo.png" height="200" align="right" style="padding:12px"/>] ### .left[[inSileco: codes, ideas and methods in ecology](https://insileco.github.io/)] --- name: title layout: true class: center, middle, style1 --- # DO BIOTIC INTERACTIONS IMPACT GEOGRAPHIC RANGE LIMITS OF SPECIES? <br> ### [Kevin Cazelles](http://kevincazelles.fr), [Universty of Guelph](https://www.uoguelph.ca/) [](https://github.com/KevCaz/seminarInteractionsDistributions) <br/> ### Département de biologie, Université de Sherbrooke #### October, 12<sup>th</sup> 2018 --- name: style2 layout: true class: center middle, style2 --- # INTRODUCTION <hr/> ### Biogeography and biotic interactions --- name: style1 layout: true class: style1 --- # Biogeography: distributions <hr/> .center[] --- # Biogeography: distributions <hr/> .center[] --- # Biogeography: processes <hr/> .center[] --- # Individuals <hr/> .center[] --- # Individuals to population <hr/> .center[] ??? Interaction at this scale that one can actually envision interactions --- # Population to metapopulation <hr/> .center[] ??? Interaction at this scale that one can actually envision interactions --- # Populations to metapopulations <hr/> .center[] ??? Interaction at this scale that one can actually envision interactions --- # Metapopulations to metacommunities <hr/> .center[] ??? Interaction at this scale that one can actually envision interactions --- # Metacommunities through time <hr/> .center[] ??? We scaled up in space Let's sclaed up in time Lots of process to take into account ! That explain why species are here... Lot's of processes le'ts recap --- # Metacommunities through time <hr/> .center[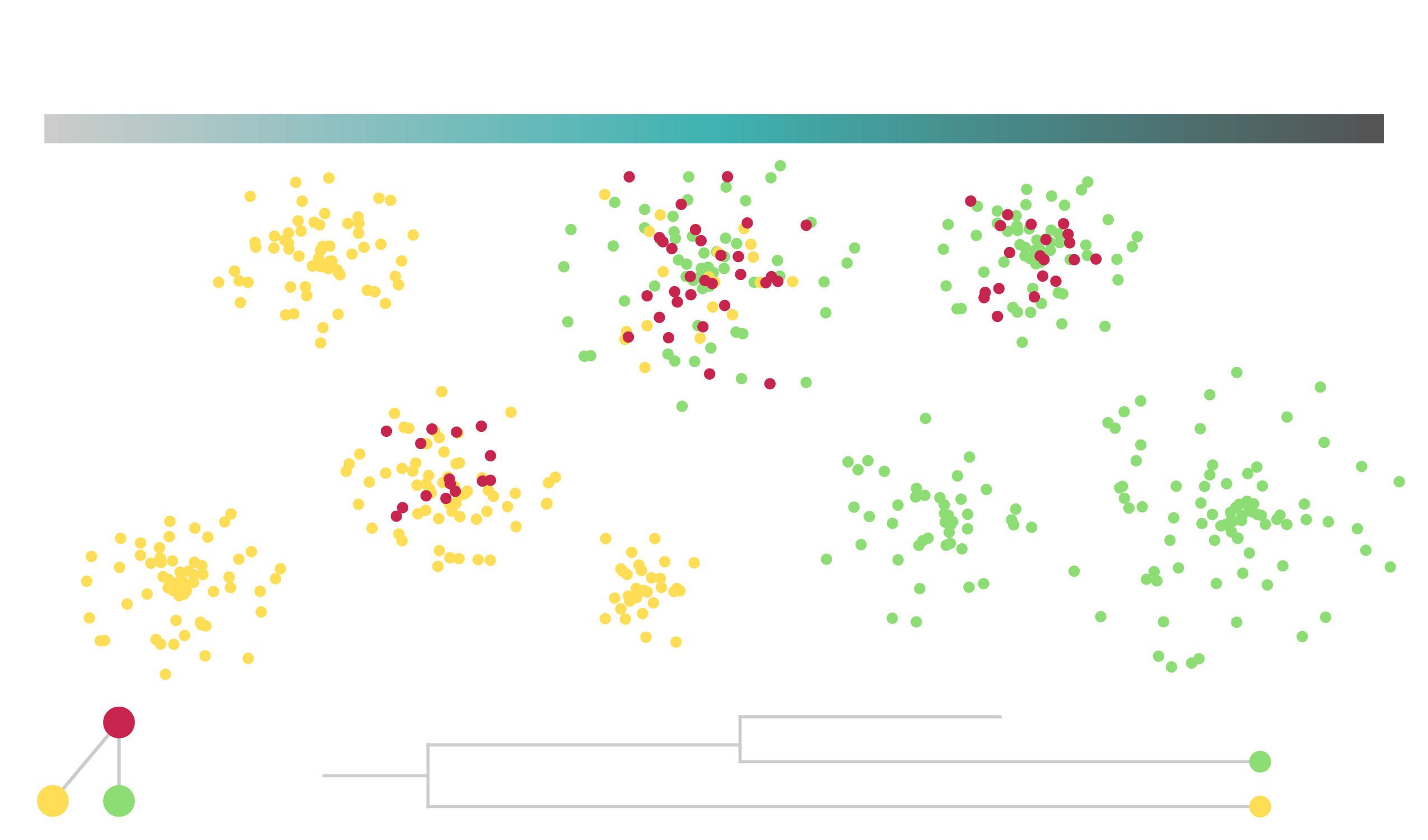] --- # Metacommunities through time <hr/> .center[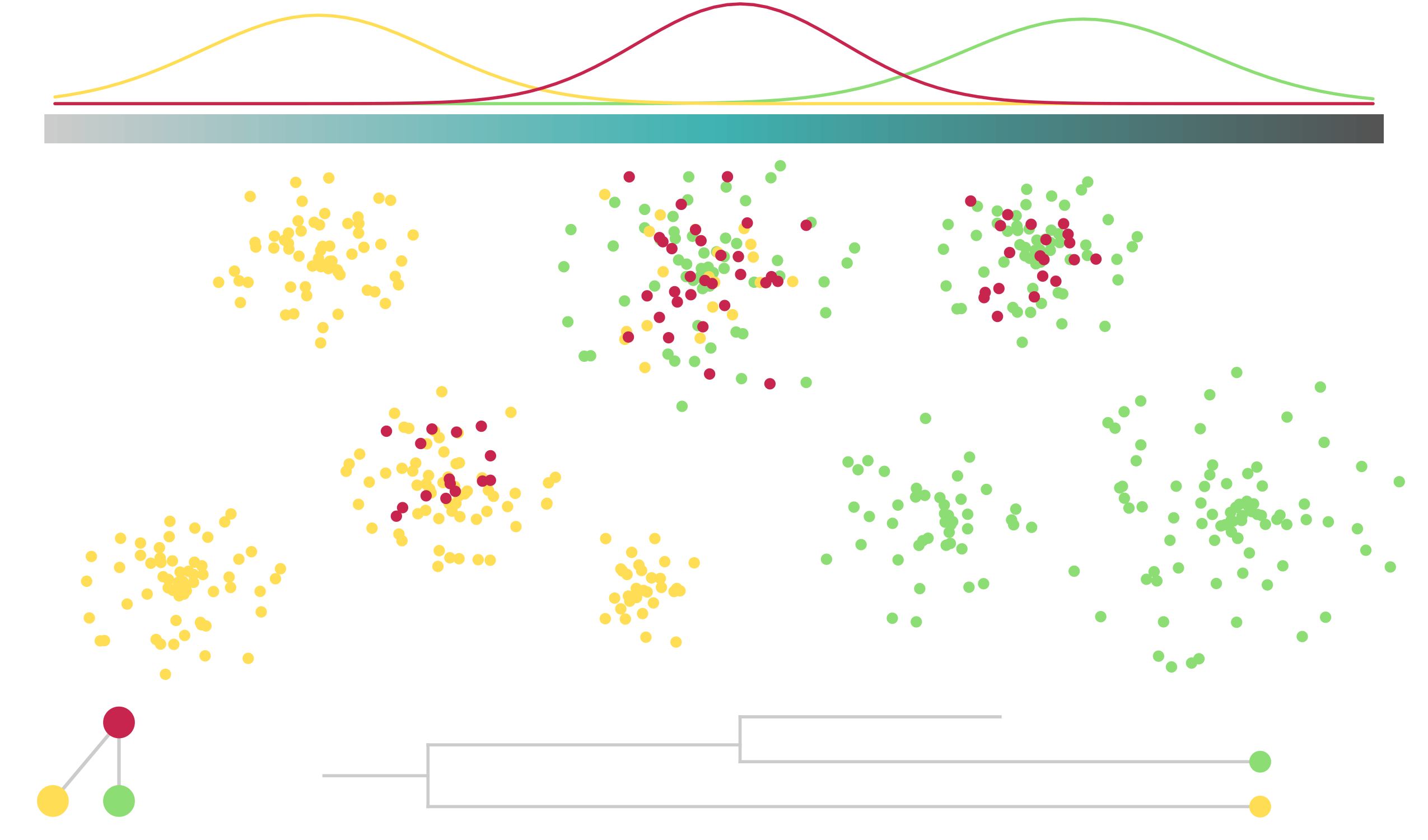] --- # Biogeography: processes <hr/><br/> ## 1. Abiotic variables -- ## 2. Dispersion -- ## 3. Ecological interactions -- ## 4. Historical factors -- <br/> # .center[[INTERDEPENDENT]()] --- # Biogeography: questions <hr/><br/> -- ## How processes generate distributions? ## How to infer processes from distributions? --- # Biogeography: challenge <hr/> .center[] --- # Biogeography: challenge <hr/> .center[] --- # Biogeography: challenge <hr/> .center[] --- # Species Distribution Models (SDMs) <hr/> <br/> .center[] --- # Species Distribution Models (SDMs) <hr/> <br/> .center[] --- # Predicting species distributions <hr/> .center[] --- # Predicting species distributions <hr/> .center[] --- # Species Distribution Models (SDMs) <hr/> <br/> .center[] --- # Species Distribution Models (SDMs) <hr/> <br/> .center[] --- # Predicting species distributions <hr/> .center[] --- # Predicting species distributions <hr/> ## Species are assumed independent -- ###  Davis *et al*, 1998, *Nature* -- <br/> ## Joint Species Distribution Models (JSDMs) ###  Clark *et al*, 2014, *Ecological Applications* ###  Pollock *et al*, 2014, *Methods in Ecology and Evolution* ###  Zurell *et al*, 2018, *Ecography* --- # Predicting species distributions <hr/><br> -- ## Theory ###  Holt, *Toward a Trophic Island Biogeography*, 2009. [](https://press.princeton.edu/titles/9096.html) ###  Gravel *et al.*, *Ecology Letters*, 2011.[]((http://onlinelibrary.wiley.com/doi/10.1111/j.1461-0248.2011.01667.x/abstract) ###  Massol *et al.*, *Advances in Ecological Research*, 2017.[]((https://doi.org/10.1016/bs.aecr.2016.10.004) --- # Biogeography: challenge <hr/> .center[] --- # Biogeography: challenge <hr/> .center[] --- # Questions <hr/><br/> ### How interactions influence species distributions? ### How to infer the role of biotic interactions from co-occurrence data? <br/><br/> -- 1. How to integrate biotic interactions into SDMs? -- 2. How the properties of ecological networks influence co-occurrence? -- 3. Can we infer interactions from co-occurrence data? ??? des why / des hypothèses et des how --- name: style2 layout: true class: center, middle, style2 --- # THEORY OF ISLAND BIOGEOGRAPHY AND BIOTIC INTERACTIONS <hr /> ### How to integrate biotic interactions into SDMs? --- name: style1 layout: true class: style1 --- # MacArthur et Wilson (1963, 1967) <hr/> --- # MacArthur et Wilson (1963, 1967) <hr/><br/> .center[] --- # MacArthur et Wilson (1963, 1967) <hr/><br/> .center[] --- # MacArthur et Wilson (1963, 1967) <hr/><br/> .center[] --- # MacArthur et Wilson (1963, 1967) <hr/><br/> .center[] --- # MacArthur et Wilson (1963, 1967) <hr/><br/> .center[] --- # MacArthur et Wilson (1963, 1967) <hr/><br/> .center[] --- # MacArthur et Wilson (1963, 1967) <hr/><br/> .center[] --- # Model properties <hr/><br/> ### 1. Colonisation/extinction dynamics -- <br/> ### 2. Equilibrium -- <br/> ### 3. Elegant, easy to build on it -- <br/> ### 4. Biotic and abiotic constraints are missing ??? ça marche bien... et très utilisé --- # Releasing the assumption of independence <hr/><br/> -- ## Gravel *et al*, 2011, *Ecology Letters* [](http://onlinelibrary.wiley.com/doi/10.1111/j.1461-0248.2011.01667.x/abstract) -- ### 1. Island without prey = no predator colonisation ### 2. Predator without prey = extinction <br/> -- ## Colonisation and extinction depend on the local community <br/> ??? première étape pour lever l'hypothèse. --- # What did I do? <hr/><br/> .center[] --- # What did I do? <hr/><br/> .center[] --- # What did I do? <hr/><br/> .center[] --- # What did I do? <hr/><br/> .center[] --- # What did I do? <hr/><br/> .center[] --- # What did I do? <hr/><br/> .center[] --- # What did I do? <hr/><br/> .center[] --- # What did I do? <hr/><br/> .center[] --- # Results: the classical theory revisited <hr/><br/> .center[] --- # Results: the classical theory revisited <hr/><br/> .center[] --- # Results: the classical theory revisited <hr/><br/> .center[] --- # Results: the classical theory revisited <hr/><br/> .center[] --- # Results: the classical theory revisited <hr/><br/> .center[] --- # Conclusion and limits <hr/><br> ### For `n` species, a network `W`, a set of abiotic variables `E`, -- <br/> ### Community `\(C_k = \{0, 1, 0, 0, ..., 1, 0\}\)` -- ## `\(P(C_{k,t+1} | C_{l, t}) = f(W, E)\)` -- <br/> ### .right[[Cazelles *et al.*, *Ecography* (2016).](http://doi.org/10.1111/ecog.01714)] --- # Conclusion and limits <hr/><br> ### Community-based biogeography: more than occurrence and co-occurrence <br/> -- ### `\(n\)` species `\(2^n\)` potential communities <br/> -- ### [What is wrong with co-occurrence data for species embedded in a network?]() --- name: style2 layout: true class: center, middle, style2 --- # CO-OCCURRENCE AND ECOLOGICAL NETWORKS <hr /> ### How the properties of ecological networks influence co-occurrence? --- name: style1 layout: true class: style1 --- # Co-occurrence and biotic interactions <hr/><br/> .center[] --- # Co-occurrence and biotic interactions <hr/><br/> .center[] --- # Co-occurrence and biotic interactions <hr/><br/> .center[] --- # Co-occurrence and biotic interactions <hr/><br/> .center[] --- # Co-occurrence and biotic interactions <hr/><br/> .center[] --- # Co-occurrence and biotic interactions <hr/><br/> .center[] --- # Checkerboard distribution (Diamond 1975) <hr/><br/> .center[] --- # Checkerboard distribution (Diamond 1975) <hr/><br/> .center[] --- # Checkerboard distribution (Diamond 1975) <hr/><br/> .center[] --- # What did I do? <hr/><br/> ### Checkerboard distrbution in a network context <br> -- .column[ ### Observed `vs` Expected <br> ### `\(P(X_i,X_j) - P(X_i)P(X_j)\)` ] .column[ ] --- # What did I do? <hr/><br/> .column[ ### Observed `vs` Expected <br> ### `\(P(X_i,X_j) - P(X_i)P(X_j)\)` ] .column[ `\(P(X_i,X_j) - P(X_i)P(X_j)=0\)` <br/> .center[] ] --- # What did I do? <hr/><br/> .column[ ### Observed `vs` Expected <br> ### `\(P(X_i,X_j) - P(X_i)P(X_j)\)` ] .column[ `\(P(X_i,X_j) - P(X_i)P(X_j)<0\)` <br/> .center[] ] --- # What did I do? <hr/><br/> .column[ ### Observed `vs` Expected <br> ### `\(P(X_i,X_j) - P(X_i)P(X_j)\)` ] .column[ `\(P(X_i,X_j) - P(X_i)P(X_j)>0\)` <br/> .center[] ] --- # What did I do? <hr/><br/> .column[ ### Observed `vs` Expected <br> ### `\(P(X_i,X_j) - P(X_i)P(X_j)\)` <br/> ### Simulated co-occurrence (trophic interactions) ] .column[ <br/> <br/> .center[] ] --- # What did I do? <hr/><br/> .column[ ### Observed `vs` Expected <br> ### `\(P(X_i,X_j) - P(X_i)P(X_j)\)` <br/> ### Simulated co-occurrence (trophic interactions) <br/> ### Simulated networks ] .column[ <br/> .center[] ] --- # Co-occurrence and shortest path <hr/><br/> .center[] --- # Co-occurrence and shortest path <hr/><br/> .center[] --- # Co-occurrence and shortest path <hr/><br/> .center[] --- # Co-occurrence and shortest path <hr/><br/> .center[] --- # Co-occurrence and degree <hr/><br/> .center[] --- # Co-occurrence and degree <hr/><br/> .center[] --- # Co-occurrence and degree <hr/><br/> .center[] --- # Co-occurrence and degree <hr/><br/> .center[] --- # Conclusion and limits <hr/><br/> ## Shortest path <i class="fa fa-long-arrow-up" aria-hidden="true"></i> `\( \Longrightarrow \)` detection <i class="fa fa-long-arrow-down" aria-hidden="true"></i> -- ## Degree <i class="fa fa-long-arrow-up" aria-hidden="true"></i> `\( \Longrightarrow \)` detection <i class="fa fa-long-arrow-down" aria-hidden="true"></i> -- <br/><br/><br/> ### .right[[Cazelles *et al.*, *Theoretical Ecology* (2016).](http://doi.org/10.1007/s12080-015-0281-9)] --- # Correlations? <br> .center[] --- # Correlations? <br> .center[] --- # Correlations? <br> .center[] --- # Correlations? <br> .center[] --- # Conclusion and limits <hr/><br/> ## Shortest path <i class="fa fa-long-arrow-up" aria-hidden="true"></i> `\( \Longrightarrow \)` detection <i class="fa fa-long-arrow-down" aria-hidden="true"></i> ## Degree <i class="fa fa-long-arrow-up" aria-hidden="true"></i> `\( \Longrightarrow \)` detection <i class="fa fa-long-arrow-down" aria-hidden="true"></i> -- <br/> ## Abiotic variables? -- ## Empirical data? --- name: style2 layout: true class: center, middle, style2 --- # DEALING WITH REAL DATA SETS <hr /> ### Can we infer interactions from co-occurrence data? --- name: style1 layout: true class: style1 --- # Abiotic variables <hr/><br/> .center[] --- # Abiotic variables <hr/><br/> .center[] --- # Integrating abiotic variables (E) <hr/><br/> # `\(P(X_{i,E},X_{j,E})\)` `vs` `\(P(X_{i,E})P(X_{j,E})\)` --- # Integrating abiotic variables (E) <hr/><br/> .center[] --- # Integrating abiotic variables (E) <hr/><br/> .center[] --- # What did I do? <hr/><br/> ### 4 data sets, **know interactions** -- <br/> ### `\(P(X_{i,E},X_{j,E})\)` `vs` `\(P(X_{i,E})P(X_{j,E})\)` -- <br/> ### SDMs to assign presence probabilities given abiotic context -- <br/> ### Detection <i class="fa fa-long-arrow-down" aria-hidden="true"></i> once abiotic variables integrated --- # Kopelke *et al.*, *Ecology* (2017) <hr/> .column[ .center[] ] .column[ .center[] ] ??? mettre les noms --- # Kopelke *et al.*, *Ecology* (2017) <hr/> .column[ .center[] ] .column[ .center[] ] ??? symphyte / ovipositeur --- # Three trophic levels network <hr/><br/> # Willow <i class="fa fa-long-arrow-right" aria-hidden="true"></i> Galler <i class="fa fa-long-arrow-right" aria-hidden="true"></i> Parasitoids # 52 <i class="fa fa-long-arrow-right" aria-hidden="true"></i> 96 <i class="fa fa-long-arrow-right" aria-hidden="true"></i> 126 species -- <br/> # Willow <i class="fa fa-long-arrow-right" aria-hidden="true"></i> Galler -- # Galler <i class="fa fa-long-arrow-right" aria-hidden="true"></i> Parasitoids --- # Willow <i class="fa fa-long-arrow-right" aria-hidden="true"></i> Galler - Shortest path <hr/><br/>  --- # Willow <i class="fa fa-long-arrow-right" aria-hidden="true"></i> Galler - Shortest path <hr/><br/>  --- # Willow <i class="fa fa-long-arrow-right" aria-hidden="true"></i> Galler - Shortest path <hr/><br/>  --- # Willow <i class="fa fa-long-arrow-right" aria-hidden="true"></i> Galler - Shortest path <hr/><br/>  --- # Willow <i class="fa fa-long-arrow-right" aria-hidden="true"></i> Galler - Shortest path <hr/><br/>  --- # Willow <i class="fa fa-long-arrow-right" aria-hidden="true"></i> Galler - `\(P(X_{i,E},X_{j,E})\)` <hr/><br/>  --- # Galler <i class="fa fa-long-arrow-right" aria-hidden="true"></i> Parasitoids - `\(P(X_{i,E},X_{j,E})\)` <hr/><br/>  --- # Co-occurrence and shortest path <hr/><br/> .center[] --- # Willow <i class="fa fa-long-arrow-right" aria-hidden="true"></i> Galler `vs` Galler <i class="fa fa-long-arrow-right" aria-hidden="true"></i> Parasitoids <hr/> .center[] .center[] --- # Willow <i class="fa fa-long-arrow-right" aria-hidden="true"></i> Galler `vs` Galler <i class="fa fa-long-arrow-right" aria-hidden="true"></i> Parasitoids <hr/> .center[] .center[] --- # Willow <i class="fa fa-long-arrow-right" aria-hidden="true"></i> Galler `vs` Galler <i class="fa fa-long-arrow-right" aria-hidden="true"></i> Parasitoids <hr/> .center[] .center[] --- # Co-occurrence and degree <hr/><br/> .center[] --- # Conclusion <hr/><br/> ## Shortest path <i class="fa fa-long-arrow-up" aria-hidden="true"></i> `\( \Longrightarrow \)` detection <i class="fa fa-long-arrow-down" aria-hidden="true"></i> -- ## Degree <i class="fa fa-long-arrow-up" aria-hidden="true"></i> `\( \Longrightarrow \)` detection <i class="fa fa-long-arrow-down" aria-hidden="true"></i> -- ## Using empirical data sets. -- <br/><br/><br/><br/><br/> ### .right[[Cazelles *et al.*, *in prep* (2018).]()] --- name: style2 layout: true class: center, middle, style2 --- # Concluding remarks and perspectives <hr/> ### Towards better predictions? --- name: style1 layout: true class: style1 --- # Answers <hr/><br/> ## 1. How to integrate biotic interactions into distribution models? -- ### `\( \Longrightarrow \)` think at the community-scale -- ### `\( \Longrightarrow \)` build Network Distribution Models (NDMs) --- # Answers <hr/><br/> ## 2. How the properties of ecological networks influence co-occurrence? -- ### `\( \Longrightarrow \)` shortest path <i class="fa fa-long-arrow-up" aria-hidden="true"></i> `\( \Longrightarrow \)` detection <i class="fa fa-long-arrow-down" aria-hidden="true"></i> ### `\( \Longrightarrow \)` degree <i class="fa fa-long-arrow-up" aria-hidden="true"></i> `\( \Longrightarrow \)` detection <i class="fa fa-long-arrow-down" aria-hidden="true"></i> -- ### We may not need to consider the whole network --- # Answers <hr/><br/> ## 3. Can we infer interactions from co-occurrence data? -- ### `\( \Longrightarrow \)` depends on the network properties -- ### `\( \Longrightarrow \)` depends on the spatial scale (literature) ??? variations des interactions --- # Is it correct? <hr/> .center[] -- ## .center[INTERDEPENDENCY] --- # Towards mechanistic NDMs? <hr/><br/> ## Better than JSDMs? (more information) -- ## Network Distribution Models (NDMs) -- <br/> ## Network `\(W\)`, abiotic variables `\(E\)` -- <br/> ## How to predict interactions? --- # Towards mechanistic NDMs? <hr/><br/> .center[] --- # Towards mechanistic NDMs? <hr/><br/> .center[] --- # Towards mechanistic NDMs? <hr/><br/> .center[] --- # Towards mechanistic NDMs? <hr/><br/> .center[] --- # Towards mechanistic NDMs? <hr/><br/> .center[] --- # Towards mechanistic NDMs? <hr/><br/> ### 1. **Building a biogeography of ecological networks** <br>  [Galiana, N., *et al.* (2018). The spatial scaling of species interaction networks. *Nature Ecology & Evolution*.](https://www.nature.com/articles/s41559-018-0517-3) <br> -- ### 2. **[Building an energetic framework](https://kevcaz.github.io/talkBES2017/#1)** --- name: style2 layout: true class: center, middle, style2 --- # THANK YOU <hr /> ### Merci --- name: style2 layout: true class: class: center, middle, style2 --- ## What am I doing these days? <hr /> ### Science, what else? --- name: style1 layout: true --- # Homogenization <hr><br>  --- # Rewiring + seasonality in food webs <hr><br><br> ### [Nature rewires in a changing world](https://peerj.com/preprints/27187/) *Nature Eco Evo* --- # Iscoscape: the push for provenance <hr><br><br> ### [Fighting noise with dimensionality](https://kevcaz.github.io/fightingNoise/#1) --- name: style2 layout: true class: center, middle, style2 --- # THANK YOU <hr /> ### Merci --- name: style1 layout: true class: center, style1 --- --- # Urgently needed <hr/><br/> ## Interactions ++ <i class="fa fa-long-arrow-right" aria-hidden="true"></i> predictions easier ### Berlow *et al*, 2009, *PNAS* -- <br/> ## WWF, Living Planet Report 2016 [<i class="fa fa-globe" aria-hidden="true"></i>](http://awsassets.wwf.ca/downloads/wwf_living_planet_report_2016___risk_and_resilience_in_a_new_era.pdf) > If current trends continue to 2020 > vertebrate populations may decline by an > average of 67 per cent compared to 1970. --- # Using species as predictors? <hr/><br/> .center[] --- # Using species as predictors? <hr/><br/> .center[] --- # Using species as predictors? <hr/><br/> .center[] --- # Using species as predictors? <hr/><br/> .center[] --- # Using species as predictors? <hr/><br/> .center[] --- # Using species as predictors? <hr/><br/> .center[] --- # Using species as predictors? <hr/><br/> .center[] --- ## Figure 1 <br/>  --- ## Figure 2 <br/>  --- ## Figure 3 <br/>  --- ## Figure 4 <br/>  --- ## Figure 5 <br/>  --- ## Figure 6 <br/>  --- ## Figure 7 <br/><br/><br/>  --- ## Figure 8 <br/><br/><br/>  --- ## Figure 9 <br/>  --- ## Figure 10 <br/> 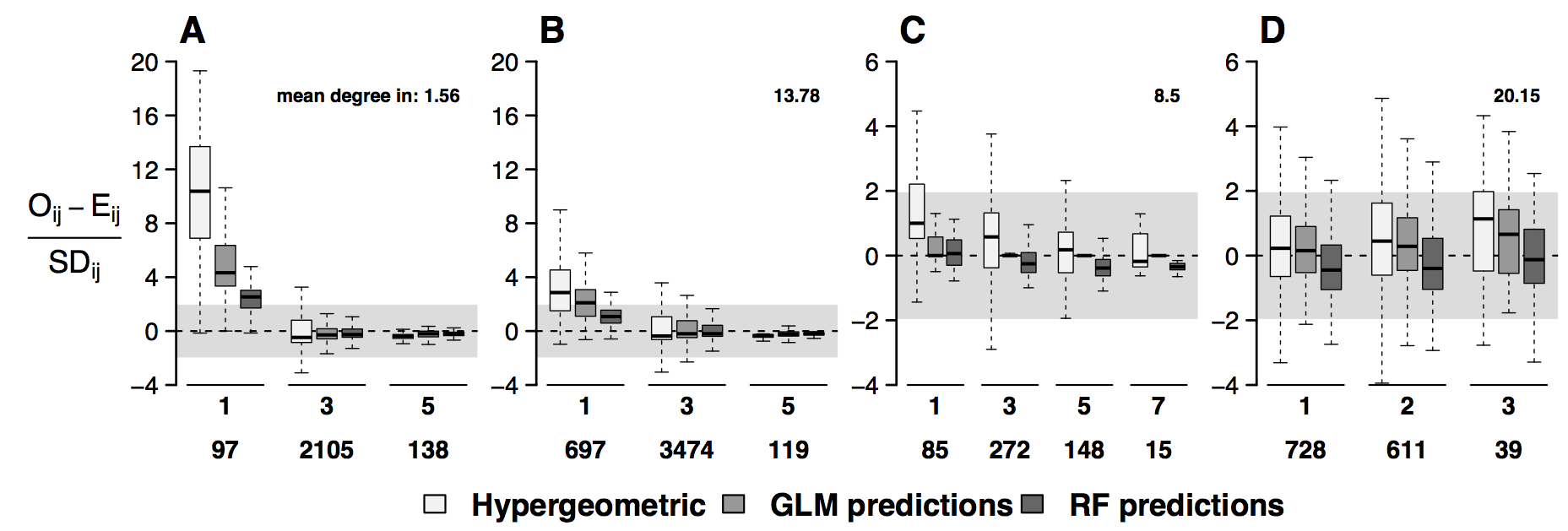 --- ## Figure 11 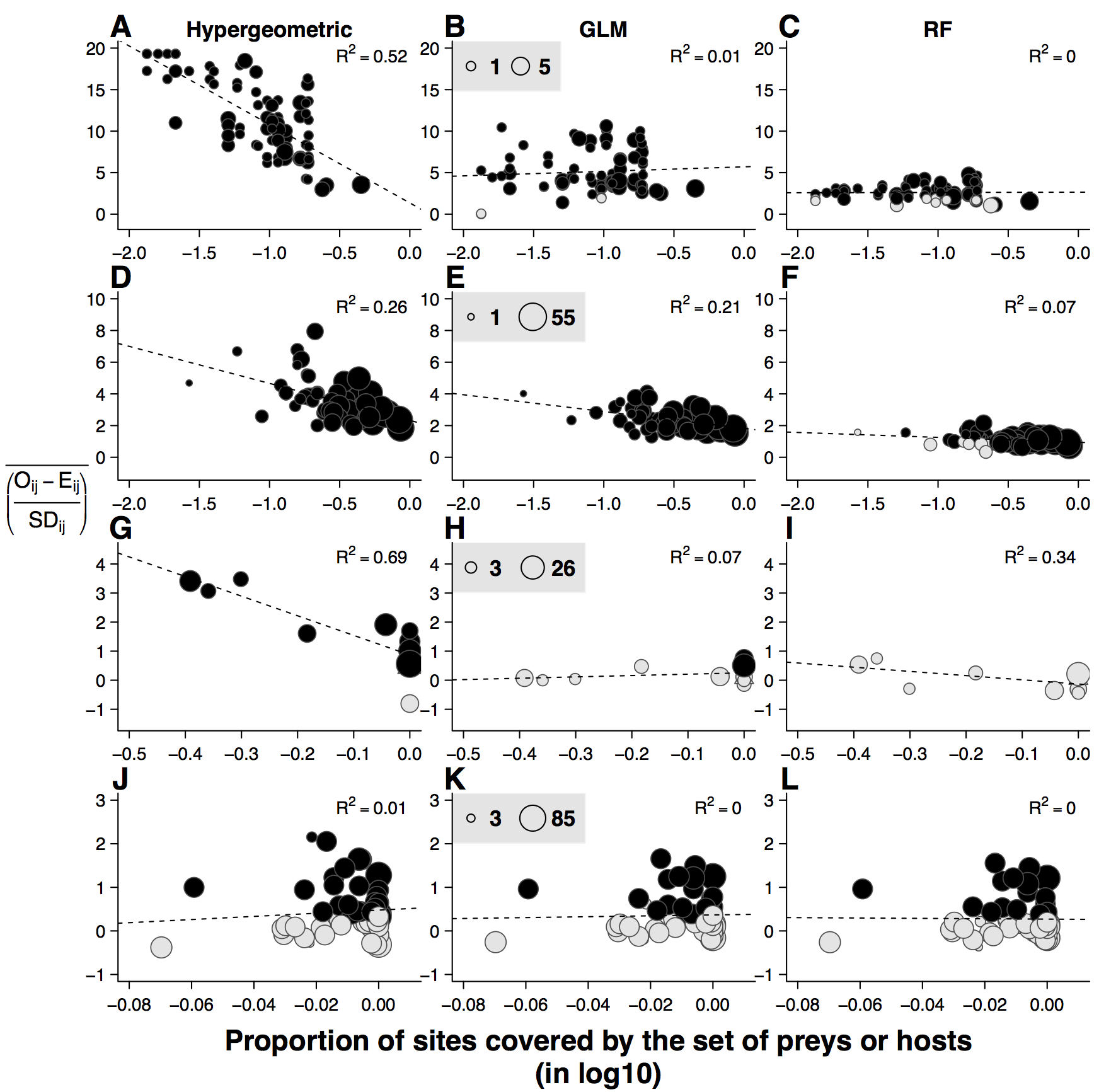 --- ## Figure 12 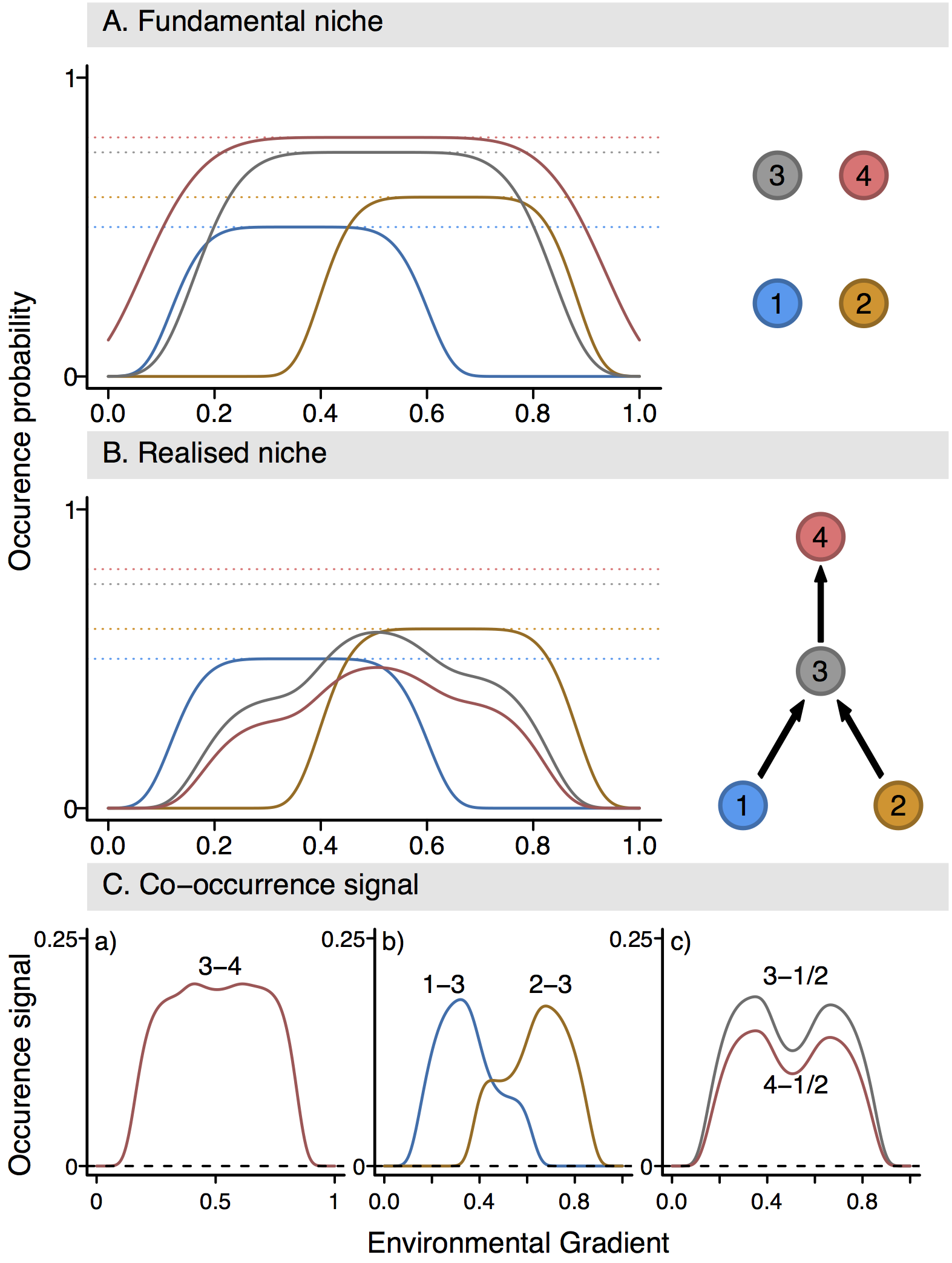 --- ## Figure 13 <br/> 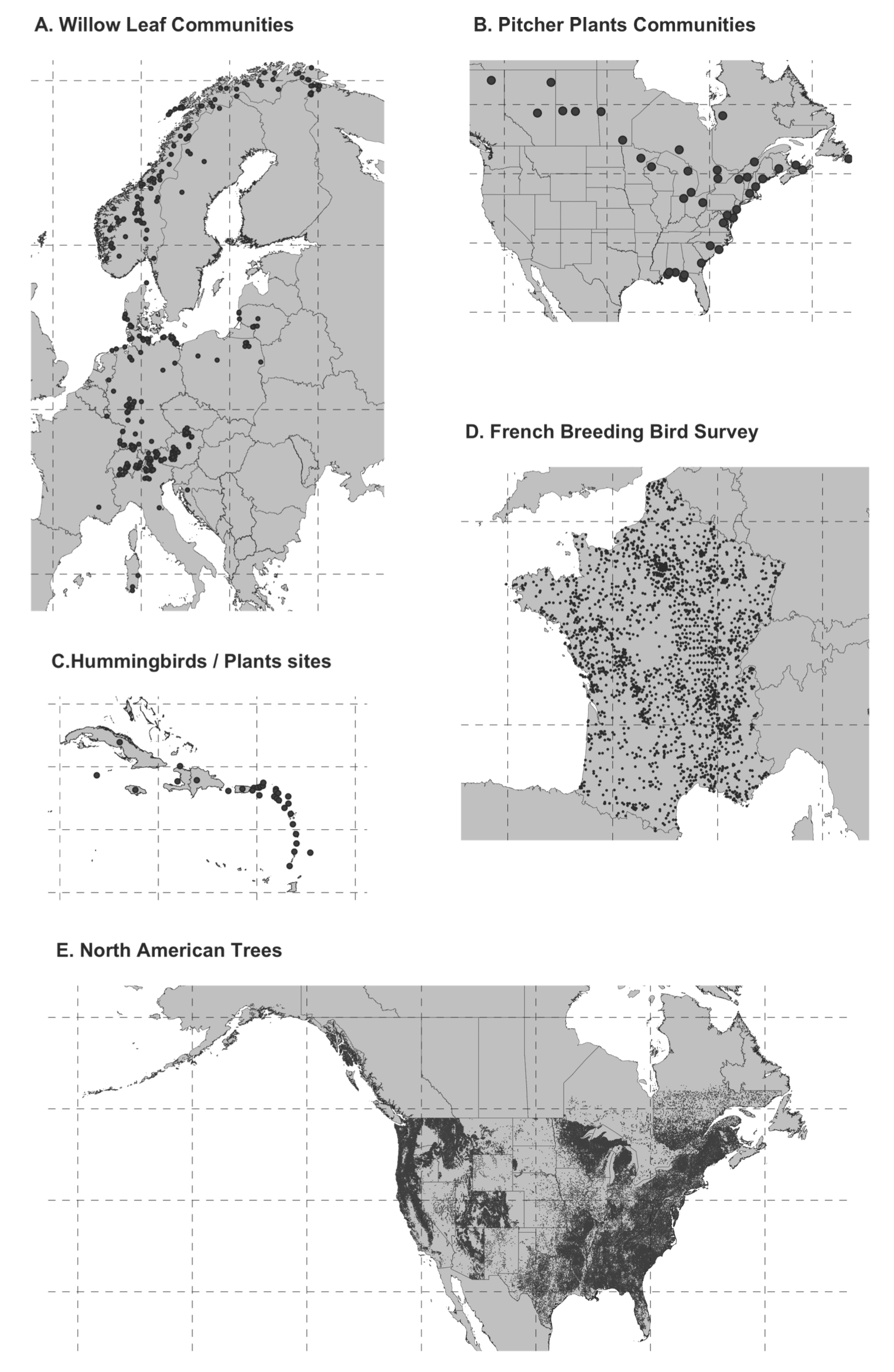 --- ## Figure 14  --- ## Figure 15 <br/> .column[] .column[] --- ## Figure 16 <br/>  --- ## Figure 17 <br/><br/><br/>  --- ## Figure 18  --- ## Figure 19  --- ## Figure 21  --- ## Figure 21 <br/>  --- ## Figure 22 <br/> .column[  ] .column[   ] --- ## Figure 23 <br/>  --- ## Figure 24 <br/>  --- ## Figure 25  --- name: style1 layout: true class: style1 --- # Phylopic <hr/><br/> - Silhouette image found on [Phylopic](http://phylopic.org/about/) - Silouhette [Ichneumonidae](http://phylopic.org/name/07b2fc56-3489-4007-904b-1a0904ea2647) ; credit Melissa Broussard. --- count:false # Salix data set <hr/> ### Species on slides 120-121: 1- Female of *Euura lapponica* ovipositing on *Salix lapponum* ; 2- leaf midrib pea gall induced by *Pontania norvegica* on *Salix borealis* ; 3- Larva of *Pontania pustulator* insid1 e opened leaf midrib bean gall on *Salix phylicifolia* ; <br/> > 641 site-visits over 29 years, and on 165,424 galls representing 96 herbivore > nodes and 52 plant nodes. The dissections and rearings yielded 42,129 natural > enemies belonging to 126 species --- name: style1 layout: true class: style1 --- count:false # Null models <hr/><br/> -- ### Since 1970 (following Diamond's paper) ### Connor et al., 2013, Ecology. <br/> -- ### Pattern-generating models based on randomization of ecological data ### Some elements are fixed, others vary randomly. --- count:false # Null models <hr/><br/> ### What would be expected in the absence of a given mechanism? <br/> ### Detection of pattern in binary presence–absence matrices. --- count: false # Ulrich & Gotelli, 2013, *Oikos* <hr/><br/> ## Test 15 indices. <br/> .center[] --- count: false # Null models <hr/><br/> > Null models of species associations should, thus, be used only to reveal the structure of co-occurrence data. ## Difficult to assign to a particular process --- count:false # MacArthur and Wilson Limits <hr/><br/> ## Releasing the assumption of a constant pool of species is a KEY -- ## Very challenging though: ### - very large pool of species ### - abondances (metapopulations) ### - evolutionnary process <br/> -- ## All solutions increase the complexity. # ABOUT ME <hr> <br> ## PhD (2012-2016) .center[] ### Dominique Gravel / Nicolas Mouquet -- <br> ## Post-doc (2017) ### Kevin S. McCann --- # A truism? <hr/><br> ### 1- Solar radiation  Autotrophs  Hetreotrophs <br> -- ### 2- Carbon source <br> -- ### 3- Water availibility <br> -- ### 4- Nutrients availibility --- # A truism? <hr/> .center[] --- # Model principle <hr/><br> .center[] --- # Model principle <hr/><br> -- ## 1- Cost of a community <br> -- ## 2- Extinctions ### - Random ###- Energy based --- # Energy cost of a network on an island <hr/><br> .center[] ### Niche model + body size to derive maintenance metabolism --- # Energy cost of a network on an island <hr/><br> .center[] --- # Energy cost of a network on an island <hr/><br> .center[] --- # Energy cost of a network on an island <hr/><br> .center[] --- # Energy cost of a network on an island <hr/><br> .center[] --- # Energy cost of a network on an island <hr/><br> .center[] --- # Cost of a community on an island <hr/> .center[] --- # Cost of a community on an island <hr/> .center[] --- # Cost of a community on an island <hr/> .center[] --- # Model <hr/><br> .center[] --- # SARs become SERs .small[[Wright, *Oikos*, 1983.](https://www.jstor.org/stable/3544109)] <hr/> .center[] --- # SARs become SERs <hr/> .center[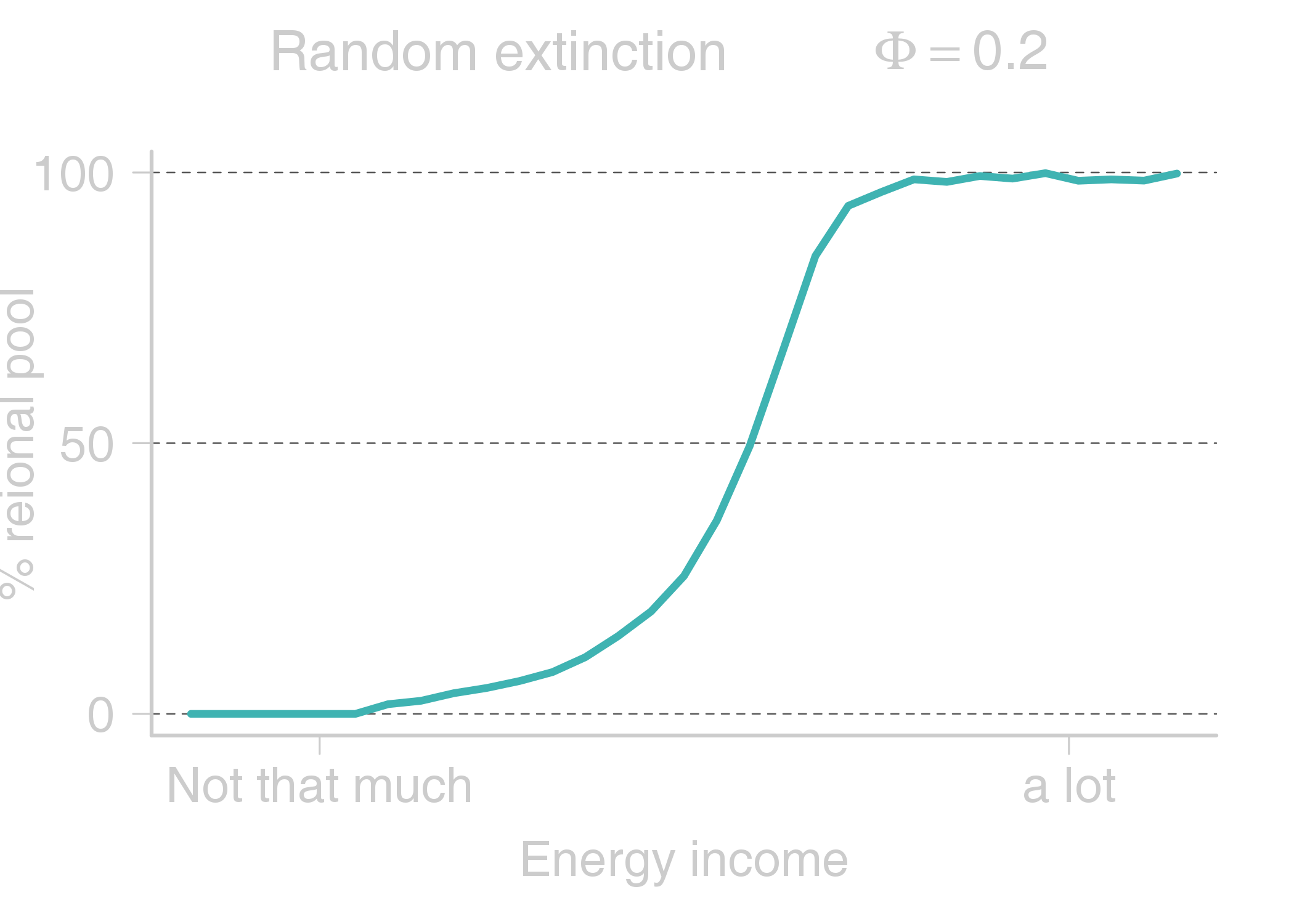] --- # SARs become SERs <hr/> .center[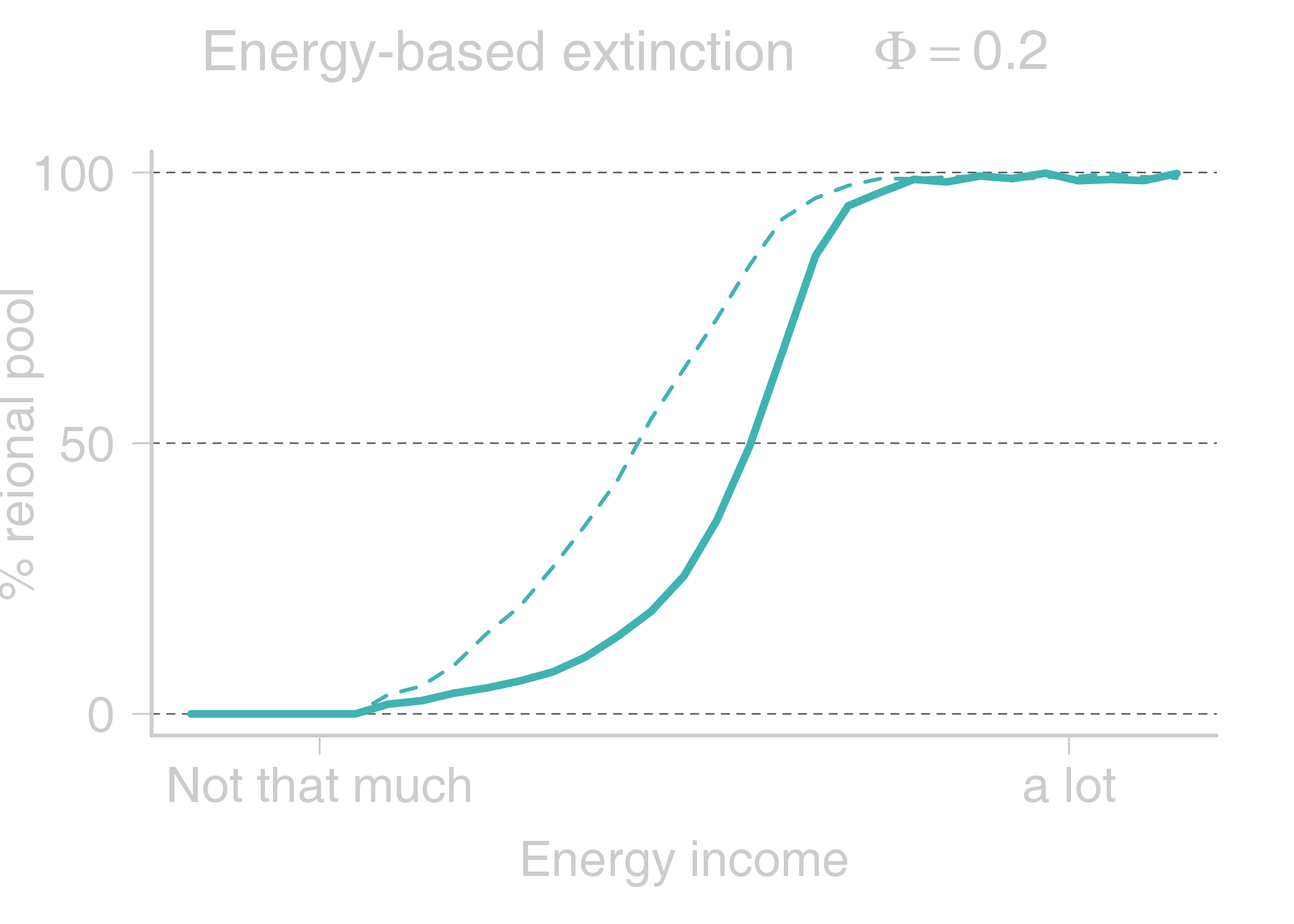] --- # SARs become SERs <hr/> .center[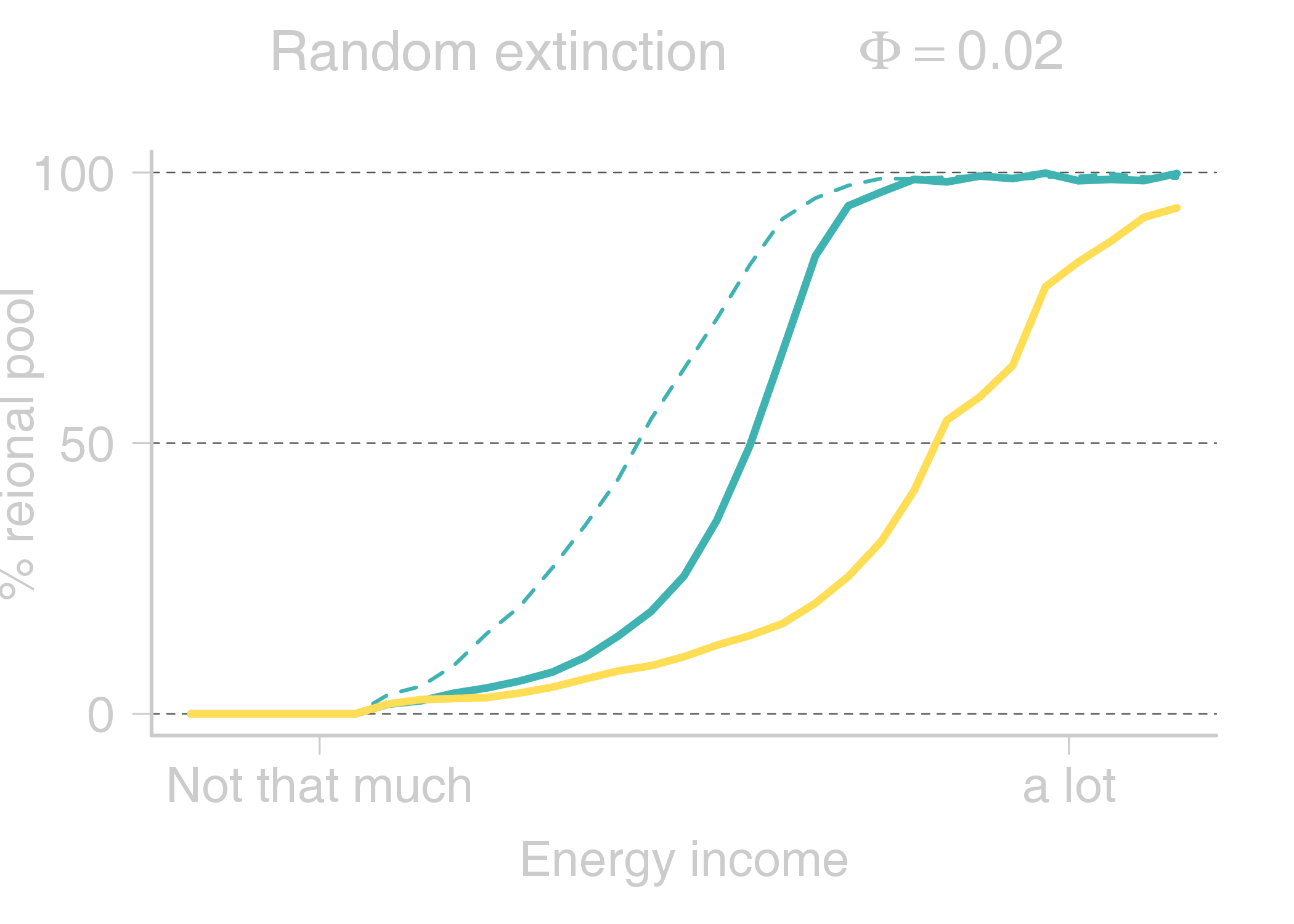] --- # SARs become SERs <hr/> .center[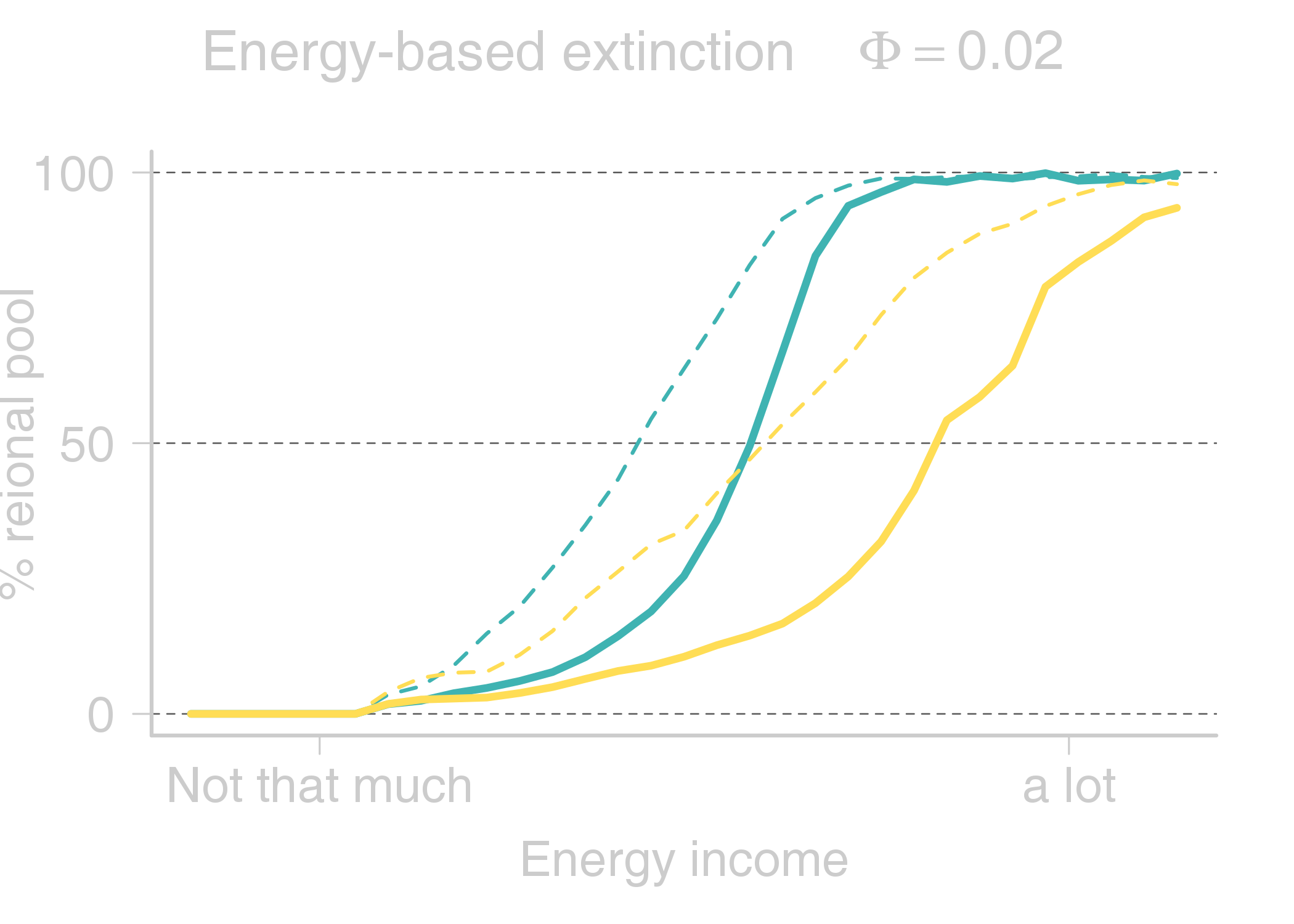] --- # Mean degree <hr/> .center[] --- # Mean degree <hr/> .center[] --- # Mean degree <hr/> .center[] --- # Mean degree <hr/> .center[] --- # Mean degree .small[[Piechnik *et al.*, *Oikos*, 2008.](https://www.jstor.org/stable/3544109)] <hr/> .center[] <!-- contrast random and non randon scenarios --> --- # An Energetic Theory of Isand Biogeography <hr/><br> -- ## - TTIB / MTE (or DEB) <br> -- ## - Better quantification <br> -- ## - Theoretical fundations of NDMs --- # New insights <hr/><br> .center[] --- # New insights <hr/> .center[]